08 Modeling Relationships
Julius-Maximilians-University Würzburg
Course: “Biostatistics”
Translational Neuroscience
Agenda
- Categorical Relationships (Chi² tests)
- Continuous Relationships (Correlations)
- Group Comparisons (t-tests & ANOVA)
Modeling Categorical Relationships
Chi² Test: General
We will first focus on modeling categorical relationships (i.e., of variables that are qualitative!).
These data are usually expressed in terms of counts.
Example: Candy colors
Bag of candy: 30 chocolates, 33 licorices, and 37 gumballs.
Is the distribution fair (i.e., 1/3rd of the bag for each candy) and the fact that there are only 30 chocolates within expectations?
What is the likelihood that the count would come out this way (or even more extreme) if the true probability of each candy type is the same?
Chi² Test: One Variable
The Chi² test checks whether observed counts differ from expected values (\(H_0\)).
\[ \chi^2 = \sum_i\frac{(observed_i - expected_i)^2}{expected_i} \]
The null hypothesis in our example is that the proportion of each type of candy is equal (1/3 or ~33.33).
If we plug in our values from above, we would calculate \(\chi^2\) like this:
\[ \chi^2 = \frac{(30 - 33.33)^2}{33.33} + \frac{(33 - 33.33)^2}{33.33} + \frac{(37 - 33.33)^2}{33.33} = 0.74 \]
Chi² Test: General
On its own, the \(\chi^2\) statistic is not interpretable - it depends on its distribution.
The shape of the \(\chi^2\) distribution depends on the degrees of freedom (much like the t distribution), which is the number of category levels \(k-1\).
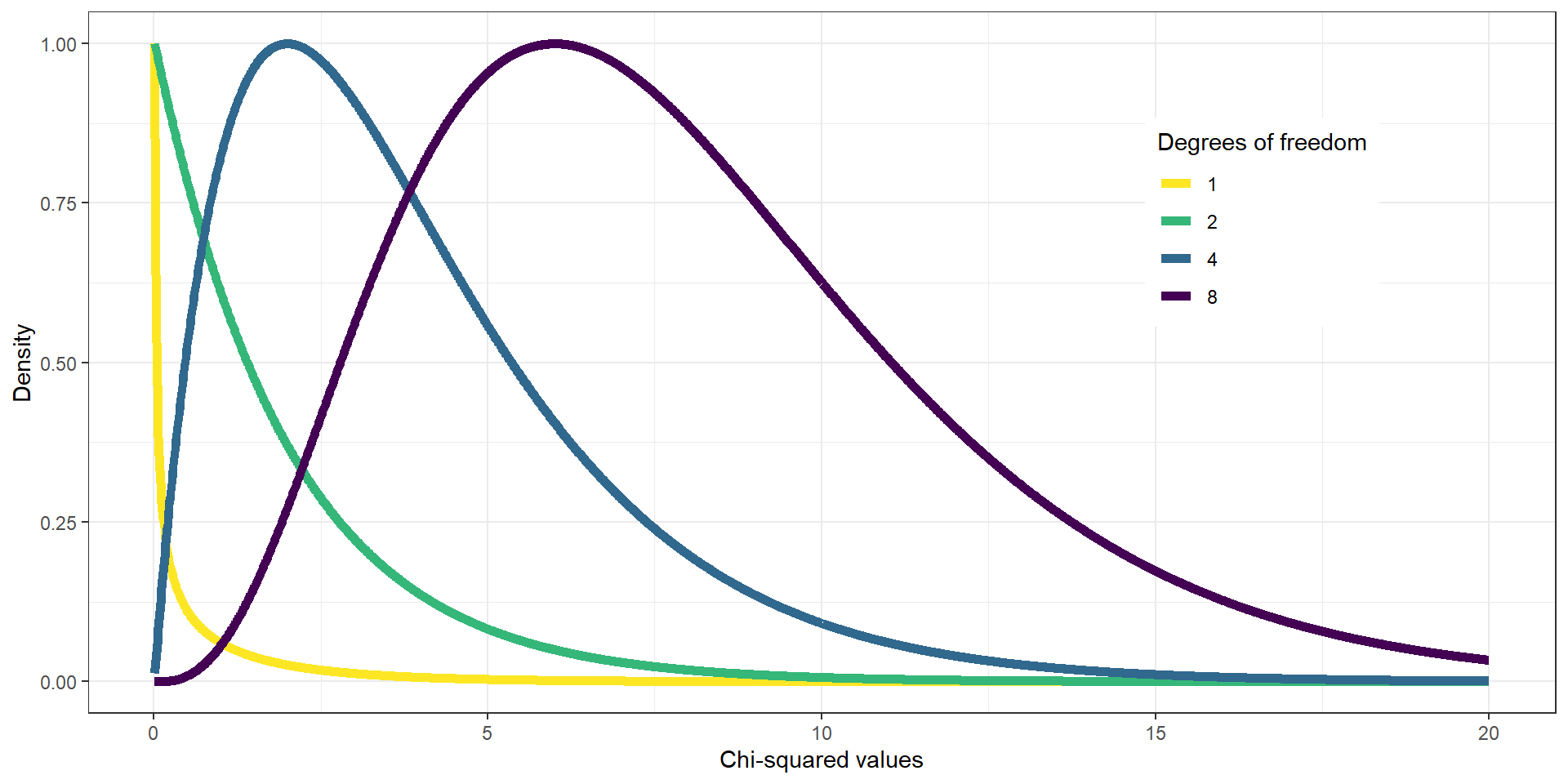
Chi2 Test: Candies
For the candy example, we use a chi-squared distribution with \(DFs = 2\) (3 candy categories minus one → green line). If we’d look at the distribution and found \(\chi^2 = .74\) on the x-axis (dashed red line), we would see that it does not fall far into the tail of the distribution but is rather in the middle. If we calculate the p-value, we’d get \(P(\chi^2 > .74) = 0.691\).
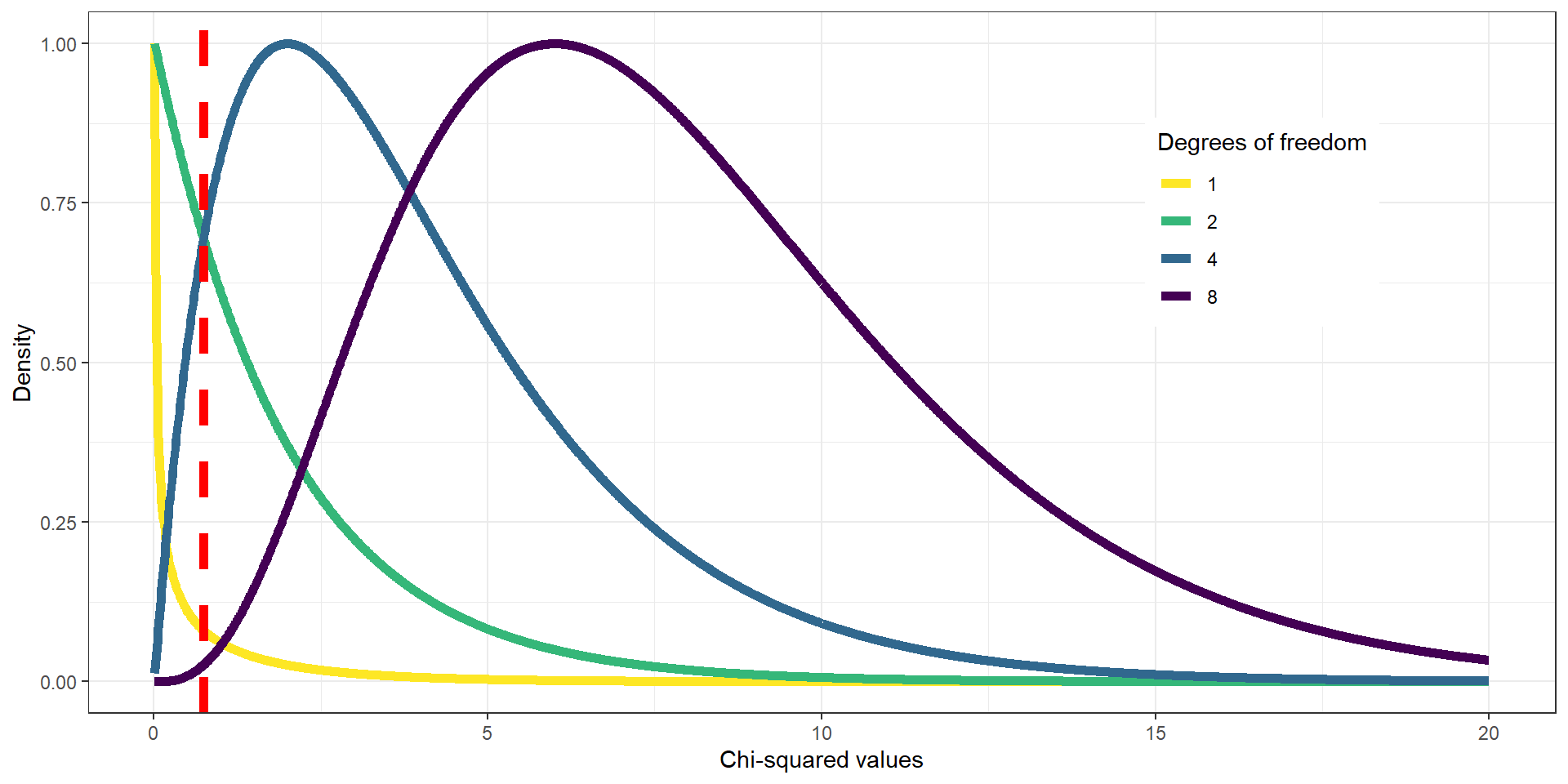
It is thus not particularly surprising to find this distribution of candies and we would not reject \(H_0\) (equal proportions).
Contingency Tables and the Two-Way Test
The \(\chi^2\) test is also used to test whether two categorical variables are related to each other.
Example: Are Black drivers more likely to be pulled over by police than white drivers?
We have two variables: Skin color (black vs white) and being pulled over (true vs. false). We can represent the data in a contingency table (remember: the count() function was helpful to do this in R):
| searched | Black | White | Black (relative) | White (relative) |
|---|---|---|---|---|
| FALSE | 36244 | 239241 | 0.1295298 | 0.8550062 |
| TRUE | 1219 | 3108 | 0.0043565 | 0.0111075 |
Contingency Tables and the Two-Way Test
If there is no relationship between skin color and being searched, the frequencies of searches would be proportional to the frequencies of skin color. This would be our expected values. We can determine them using probabilities:
| Black | White | ||
|---|---|---|---|
| Not searched | \(P(\neg S)*P(B)\) | \(P(\neg S)*P(W)\) | \(P(\neg S)\) |
| Searched | \(P(S)*P(B)\) | \(P(S)*P(W)\) | \(P(S)\) |
| \(P(B)\) | \(P(W)\) | \(100\%\) |
Remember: You can only multiply two probabilities to get their conjoint probability if they are independent. Here, we assume independence to calculate expected values and then check how much our observed values deviate from independence.
Chi² Test with Two Variables
If we compute the standardized squared difference between observed and expected values, we can sum them up to get \(\chi^2 = 828.3\)
| searched | driver_race | n | expected | stdSqDiff |
|---|---|---|---|---|
| FALSE | Black | 36244 | 36883.67 | 11.09 |
| TRUE | Black | 1219 | 579.33 | 706.31 |
| FALSE | White | 239241 | 238601.33 | 1.71 |
| TRUE | White | 3108 | 3747.67 | 109.18 |
We can then compute the p-value using a chi-squared distribution with \(DF = (levels_{var1} - 1) * (levels_{var2} - 1) = (2-1) * (2-1) = 1\)
Chi² Test with Two Variables in R
We can calculate a \(\chi^2\) test easily in R:
Pearson's Chi-squared test
data: summaryDf2wayTable
X-squared = 828.3, df = 1, p-value < 2.2e-16The results indicate that the data are highly unlikely if there was no true relationship between skin color and police searches! We would thus reject \(H_0\).
Chi² Test with Two Variables: Interpretation
Question: What is the direction of the observed relationship?
| searched | driver_race | n | expected | stdSqDiff |
|---|---|---|---|---|
| FALSE | Black | 36244 | 36883.67 | 11.09 |
| TRUE | Black | 1219 | 579.33 | 706.31 |
| FALSE | White | 239241 | 238601.33 | 1.71 |
| TRUE | White | 3108 | 3747.67 | 109.18 |
Standardized Residuals
If we want to know not only whether but also how the data differ from what we would expect under \(H_0\), we can examine the residuals of the model.
The residuals tell us for each cell how much the observed data deviates from the expected data.
To make the residuals better comparable, we will look at the standardized residuals:
\[ \text{standardized residual}_{ij} = \frac{observed_{ij} - expected_{ij}}{\sqrt{expected_{ij}}} \]
where \(i\) and \(j\) are the rows and columns respectively.
Standardized Residuals
Negative residuals indicate an observed value smaller than expected.
| searched | driver_race | Standardized residuals |
|---|---|---|
| FALSE | Black | -3.330746 |
| TRUE | Black | 26.576456 |
| FALSE | White | 1.309550 |
| TRUE | White | -10.449072 |
Odds Ratios
Alternatively, we can represent the relative likelihood of different outcomes as odds ratios:
\[ odds_{searched|black} = \frac{N(searched\cap black)}{N(\neg searched \cap black)} = \frac{1219}{36244} = 0.034 \]
\[ odds_{searched|white} = \frac{N(searched \cap white)}{N(\neg searched \cap white)} = \frac{3108}{239241} = 0.013 \]
\[ odds\ ratio = \frac{odds(searched|black)}{odds(searched|white)} = 2.59 \]
The odds of being searched are 2.59x higher for Black vs. white drivers!
Simpson’s Paradox
The Simpson’s Paradox is a great example of misleading summaries.
If we look at the baseball data below, we see that David Justice has a better batting average in every single year, but Derek Jeter has a better overall batting average:
| Player | 1995 | 1996 | 1997 | Combined | ||||
|---|---|---|---|---|---|---|---|---|
| Jeter | 12/48 | .250 | 183/582 | .314 | 190/654 | .291 | 385/1284 | .300 |
| Justice | 104/411 | .253 | 45/140 | .321 | 163/495 | .329 | 312/1046 | .298 |
How can this be?
Simpson’s Paradox: A pattern is present in the combined dataset but may be different in subsets of the data.
Happens if another (lurking) variable changes across subsets (e.g., the number of at-bats, i.e., the denominator).
Simpson’s Paradox: More intuitive
While typing on a keyboard, what is the relationship between speed (words per minute) and accuracy (% correct words)? (positive, negative, or none)
If I try to type faster, I will make more errors ⇒ speed-accuracy trade-off (negative association)
People who are better at typing usually are both faster and make fewer mistakes (positive association)
Answer: It depends
- Within a person, the relationship between speed and accuracy is negative. My own skill is limited, I can either use it for speed or accuracy.
- Between persons, the relationship is positive due to inter-individual differences in overall typing skill.
Modeling Continuous Relationships
Example: Income Inequality and Hate Crimes
We want to look at a dataset that was used for an analysis of the relationship between income inequality (Gini index) and the prevalence of hate crimes in the USA.
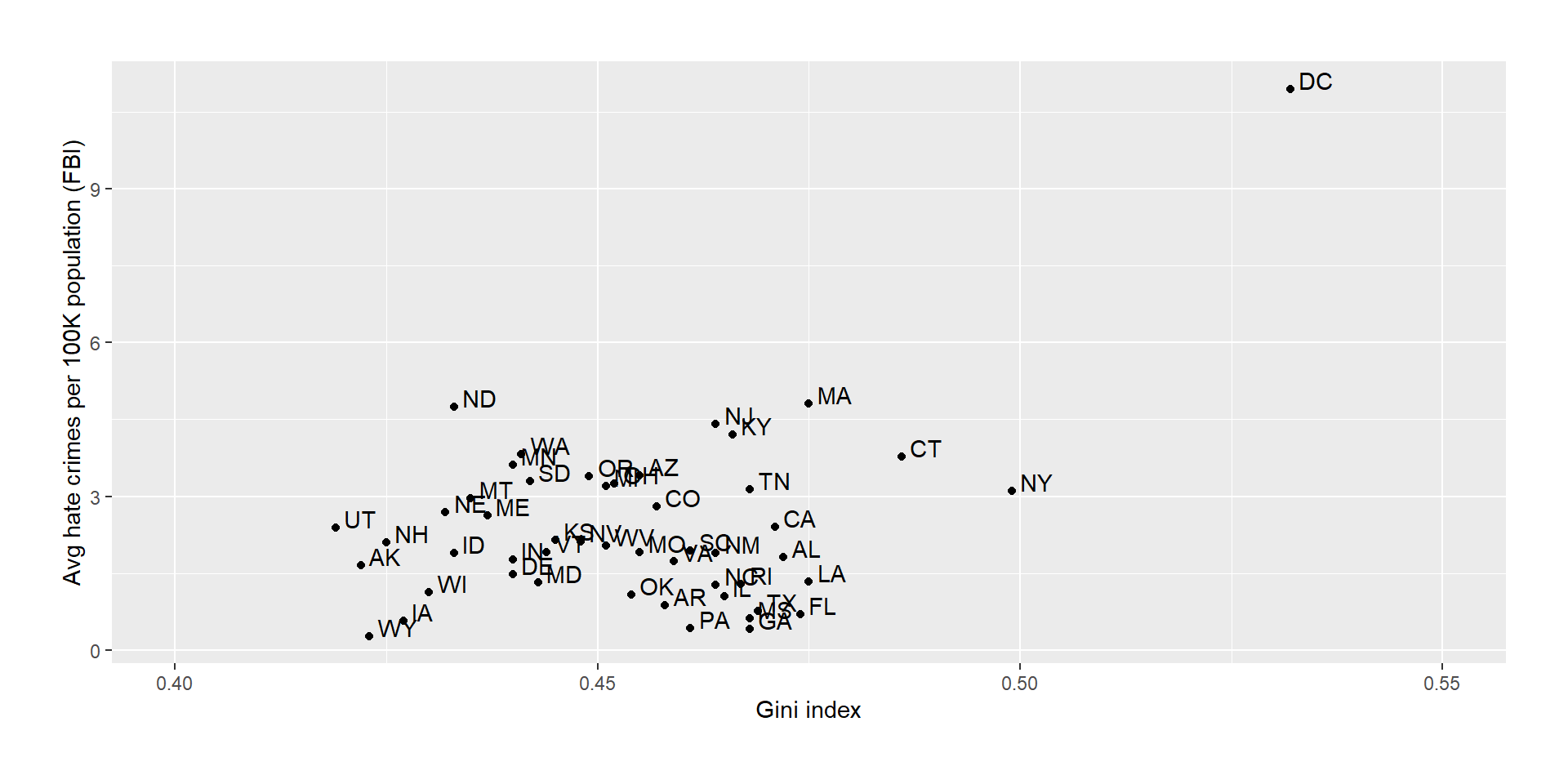
It looks like there is a positive relationship between the variables. How can we quantify this relationship?
Covariance and Correlation
Variance (single variable): \(s^2 = \frac{\sum_{i=1}^n (x_i - \bar{x})^2}{N - 1}\)
Covariance (two variables): \(covariance = \frac{\sum_{i=1}^n (x_i - \bar{x})(y_i - \bar{y})}{N - 1}\)
“Is there a relation between the deviations of two different variables (from their means) across observations?”
Covariance will be far from 0 if data points share a relationship. Positive values for same direction, negative values for opposite directions.
Covariance varies with overall level of variance in the data ⇒ hard to compare different relationships
Correlation coefficient (Pearson correlation): Scales the covariance by the standard deviations of the two variables and thus standardizes it (→ \(r\) varies between \(-1\) and \(1\) ⇒ comparability!)
\[ r = \frac{covariance}{s_xs_y} = \frac{\sum_{i=1}^n (x_i - \bar{x})(y_i - \bar{y})}{(N - 1)s_x s_y} \]
Hypothesis Testing for Correlations
The correlation between income inequality and hate crimes is \(r = .42\), which seems to be a reasonably strong (positive) relationship.
We can test whether such a relationship could occur by chance, even if there is actually no relationship. In this case, our null hypothesis is \(H_0: r = 0\).
To test whether there is a significant relationship, we can transform the \(r\) statistic into a \(t\) statistic:
\[ t_r = \frac{r\sqrt{N-2}}{\sqrt{1-r^2}} \]
Correlation in R
# perform correlation test on hate crime data
cor.test(
hateCrimes$avg_hatecrimes_per_100k_fbi,
hateCrimes$gini_index
)
Pearson's product-moment correlation
data: hateCrimes$avg_hatecrimes_per_100k_fbi and hateCrimes$gini_index
t = 3.2182, df = 48, p-value = 0.002314
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.1619097 0.6261922
sample estimates:
cor
0.4212719 The p-value is quite small, which indicates that it is quite unlikely to find an \(r\) value this high or more extreme with 48 degress of freedom. We would thus reject \(H_0: r = 0\).
Robust Correlations
In the plot, we have seen an outlier: The District of Columbia was quite different from the other data points.
The Pearson’s correlation coefficient \(r\) is highly sensitive to outliers, see this hypothetical example:
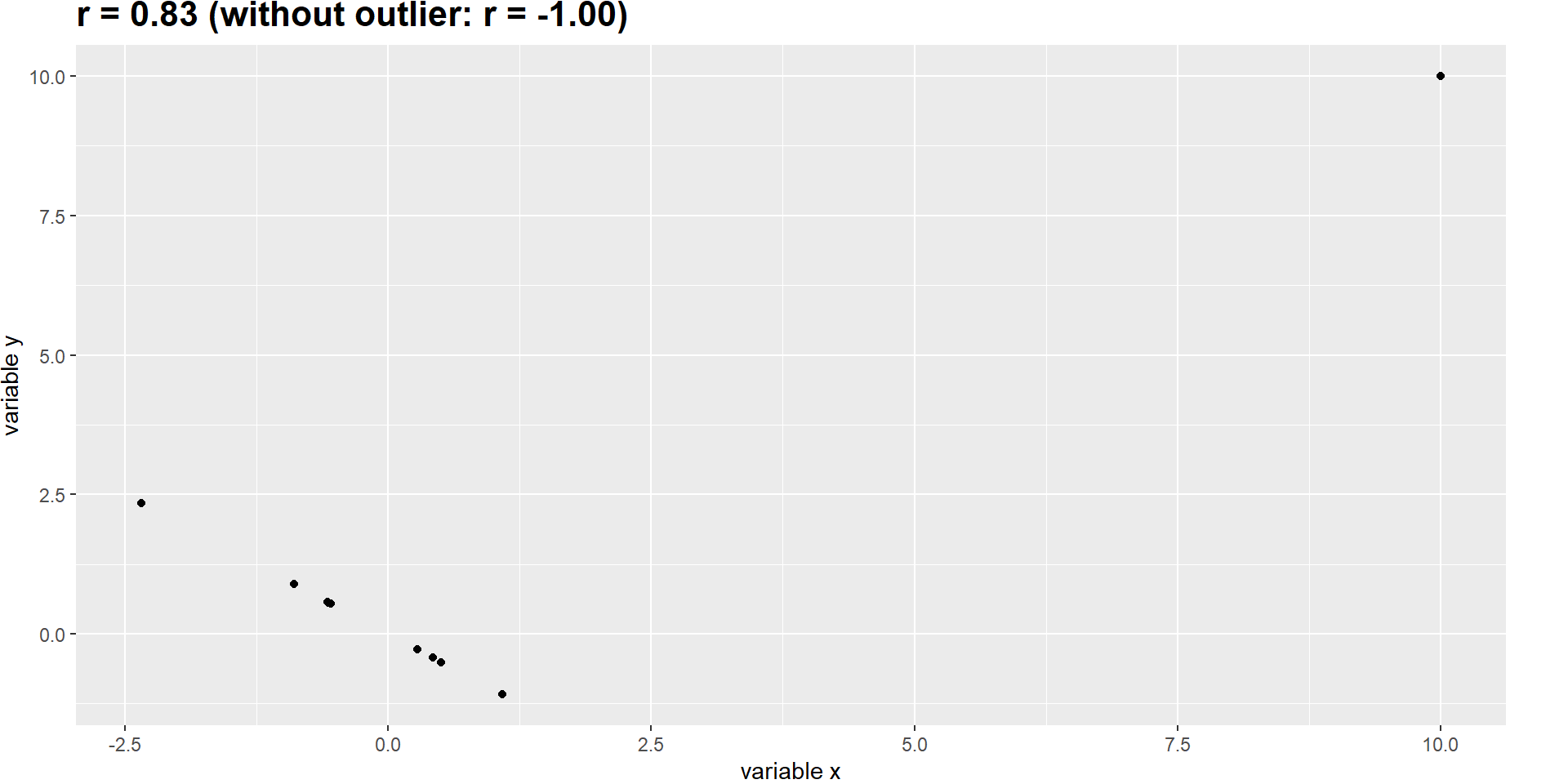
Robust Correlation 2
We can use a different correlation coefficient, which is less sensitive to outliers: Spearman correlation.
It is based on ranking (i.e., ordering) the data and using the ranks (instead of the original data) for the correlation.
Spearman's rank correlation rho
data: hateCrimes$avg_hatecrimes_per_100k_fbi and hateCrimes$gini_index
S = 20146, p-value = 0.8221
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.03261836 The correlation has now dropped from \(.42\) to \(.03\) and is no longer significant.
Of course, a ranked correlation can also obscure true effects that critically depend on the scale of the data. When in doubt, try both and discuss reasons for differences.
Correlation and Causation
Doing a well controlled, randomized experiment (RCT) is extremely helpful to gather causal evidence. However, it is not always possible or ethical to do an experiment!
Without an experiment, we can still collect observational data. However, if we correlate two observed variables, we can’t conclude that one causes the other: They (likely) share a relationship but there could also be a third variable that causes both (or even more complex causal structures).
Causal Graphs
If we have observational data, causal graphs can be helpful for interpreting causality:

Circle: observed variables
rectangle: latent (unobservable) variable
Green arrow: positive relationship,
red: negative
ExamGrade and FinishTime seem negatively related if we ignore other variables!
(“Hand in your exam early to improve your grade!”)
Knowledge = theoretical mediator
StudyTime = proxy for knowledge
If we control for StudyTime (which approximates individual knowledge), we find out that ExamGrade and FinishTime are (causally) unrelated!
Comparing Means (Group Differences)
Testing a Single Mean (One sample t-Test)
We sometimes might want to know whether a single value, the mean of a group, differs from a specific value, e.g. whether the blood pressure in the sample differs from or is bigger than 80.
We can test this using the \(t\)-test, which we have already encountered in the “Hypothesis Testing” session!
\[ t = \frac{\hat{X} - \mu}{SEM} \]
\[ SEM = \frac{\hat{\sigma}}{\sqrt{n}} \]
\(\hat{X}\) is the mean of our sample, \(\mu\) the hypothesized population mean (e.g. the value we want to test against, such as 80 for the blood pressure example).
One sample t-Test in R
One Sample t-test
data: NHANES_adult$BPDiaAve
t = -55.23, df = 4599, p-value = 1
alternative hypothesis: true mean is greater than 80
95 percent confidence interval:
69.1588 Inf
sample estimates:
mean of x
69.47239 The observed mean is actually much below 80, providing no evidence for our directional alternative hypothesis that it would be greater than 80 ⇒ the p values is (rounded to) 1.
If we had tested two-sidedly, we would have found a significant result. It may seem tempting now to create a story around an altered, non-directional alternative hypothesis, which is why it is important to preregister your hypotheses and analyses.
Comparing Two Means (Independent samples t-Test)
More often, we want to know whether there is a difference between the means of two groups.
Example: Do regular marijuana smokers watch more television?
\(H_0\) = marijuana smokers watch less or equally often TV,
\(H_A\) = marijuana smokers watch more TV.
If the observations are independent (i.e. you really have two unrelated groups), you can use a very similar formula to calculate the \(t\) statistic:
\[ t = \frac{\bar{X_1} - \bar{X_2}}{\sqrt{\frac{S_1^2}{n_1} + \frac{S_2^2}{n_2}}} \]
whereby \(\hat{X_1}\) and \(\hat{X_2}\) are the group means, \(S_1^2\) and \(S_2^2\) the group variances, and \(n_1\) and \(n_2\) the group sizes.
Independent samples t-Test in R
# t.test(NHANES_sample$TVHrsNum[NHANES_sample$RegularMarij=="Yes"],
# NHANES_sample$TVHrsNum[NHANES_sample$RegularMarij=="No"],
# alternative = "greater")
with(NHANES_sample, #only refer to data once using "with" function
t.test(TVHrsNum[RegularMarij=="Yes"],
TVHrsNum[RegularMarij=="No"],
alternative = "greater"))
Welch Two Sample t-test
data: TVHrsNum[RegularMarij == "Yes"] and TVHrsNum[RegularMarij == "No"]
t = 1.2147, df = 116.9, p-value = 0.1135
alternative hypothesis: true difference in means is greater than 0
95 percent confidence interval:
-0.09866006 Inf
sample estimates:
mean of x mean of y
2.30000 2.02963 Comparing Dependent Observations (Paired samples t-test)
If we have repeated observations of the same subject (i.e., a within-subject design), we might want to compare the same subject thus on multiple, repeated measurements.
If we want to test whether blood pressure differs between the first and second measurement session across individuals, we can use a paired \(t\)-test.
Side note: A paired \(t\)-test is equivalent to a one-sample \(t\)-test, when using the paired differences (\(X_2 - X_1\)) as \(\hat{X}\) and testing them against a value of 0 (if we expect no difference for \(H_0\)) for \(\mu\).
Paired samples t-test in R
Paired t-test
data: NHANES_sample$BPSys1 and NHANES_sample$BPSys2
t = 2.7369, df = 199, p-value = 0.006763
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
0.2850857 1.7549143
sample estimates:
mean difference
1.02 Comparing More Than Two Means
Often, we want to compare more than two means, e.g. different treatment groups or time points.
Example: Different treatments for blood pressure
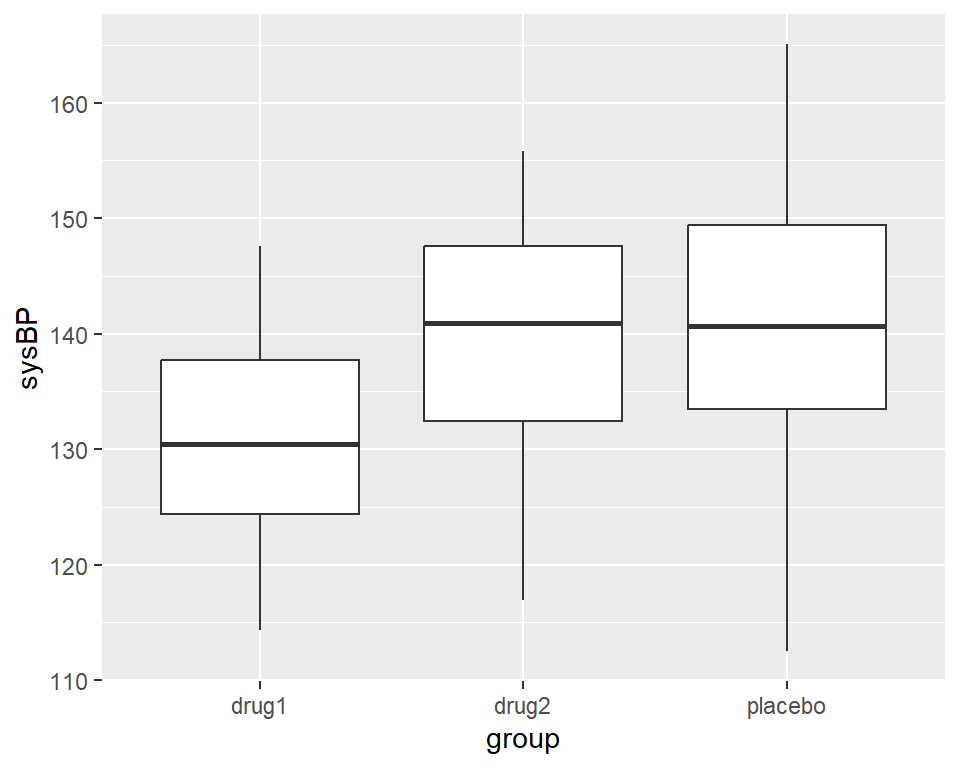
Analysis of Variance (ANOVA)
\(H_0\): all means are equal
\(H_A\): not all means are equal (e.g. at least one differs)
We can partition the variance of the data into different parts:
\(SS_{total}\) = total variance in the data
\(SS_{model}\) = Variance explained by the model
\(SS_{error}\) = Variance not explained by the model
We can use those to calculate the mean squares for the model and the error:
\(MS_{model} =\frac{SS_{model}}{df_{model}}= \frac{SS_{model}}{p-1}\) (\(p\) is the number of factor levels or groups)
\(MS_{error} = \frac{SS_{error}}{df_{error}} = \frac{SS_{error}}{N - p}\) (\(N\) is the total sample size across groups)
ANOVA in R
In R, we would run an ANOVA like this:
df <-
df %>%
mutate(group2=fct_relevel(group,c("placebo","drug1","drug2")))
# reorder the factor levels so that "placebo" is the control condition/intercept!
# test model without separate duymmies
lmResultAnovaBasic <- lm(sysBP ~ group2, data=df)
anova(lmResultAnovaBasic)Analysis of Variance Table
Response: sysBP
Df Sum Sq Mean Sq F value Pr(>F)
group2 2 2115.6 1057.81 10.714 5.83e-05 ***
Residuals 105 10366.7 98.73
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The \(F\)-statistic (also called omnibus test) actually tests our overall hypothesis of no difference between conditions.
ANOVA in R
We now know that not all groups should be considered equal. But where is the difference?
Call:
lm(formula = sysBP ~ group2, data = df)
Residuals:
Min 1Q Median 3Q Max
-29.0838 -7.7452 -0.0978 7.6872 23.4313
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 141.595 1.656 85.502 < 2e-16 ***
group2drug1 -10.237 2.342 -4.371 2.92e-05 ***
group2drug2 -2.027 2.342 -0.865 0.389
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 9.936 on 105 degrees of freedom
Multiple R-squared: 0.1695, Adjusted R-squared: 0.1537
F-statistic: 10.71 on 2 and 105 DF, p-value: 5.83e-05We can see a \(t\)-test for every drug against the placebo (remember we used fct_relevel for this). The \(t\)-tests show that drug1 (but not drug2) differs significantly from placebo.
ANOVA in R: Comments
In this simple example, we could have skipped the omnibus test (anova()) and just looked at the summary().
In practise, you will often run ANOVAs with several factors (grouping variables) that all have a multitude of factor levels (subgroups).
Think: Two groups of drugs and one placebo across four time points (before, during, after, & follow-up). This quickly creates too many possible comparisons to keep track of.
This complicates things furhter because then you can also have interactions between variables (e.g., drug1 does not work better but faster than drug2).
For these examples, it is helpful to first get an overall statement if a factor (or an interaction of factors) has an influence on the results. If this is the case, you can run the individual t-tests for this factor to get a more detailed look.
Thanks!
Learning objectives:
What are contingency tables and how do we use \(\chi^2\)-tests to assess a significant relationship between categorical variables?
Be able to describe what a correlation coefficient is as well as compute and interpret it.
Know what a \(t\)-test & ANOVA is and how to compute and interpret them.
Next:
Practical exercises in R!