1.3 Data Visualization
Dr. Mario Reutter & Juli Nagel
(slides adapted from Dr. Lea Hildebrandt)
Data Viz
Why should we visualize our data?
check whether data make sense (unusual observations?)
honestly and comprehensively present the data
(visually) check whether data fits the assumptions of statistical tests
It’s fun! (And plots are probably the most important information in papers!)
ggplot
We will use a package called ggplot2 (part of the tidyverse). ggplot2 is a very versatile package that allows us to make beautiful, publication-ready(-ish) figures.
ggplot2 follows the “grammar of graphics” by Leland Wilkinson, a formal guide to visualization principles. A core feature are the layers each plot consists of. The main function to “start” plotting is ggplot() - we will then add layers of data and layers to tweak the appearance.
Layers of a ggplot
Activity 1: Set Up
Within your precision workshop project in RStudio, create a new script called
DataVisualisation1.R.Make sure you have the following two files downloaded into your project folder (we already used them in Intro to
Rpresentation):ahi-cesd.csvandparticipant-info.csv.Copy and run the code below to load the
tidyversepackage and the data files:
Activity 1: Set Up
- Run the following code to combine both files and select our variables of interest:
all_dat <-
dat %>%
inner_join(
pinfo, # combine dat with pinfo
by = c("id", "intervention") # common variables that tell R which data belongs together
) %>%
arrange(id, occasion) #joining messes up the order of the data frame => arrange again
# we throw out several variables even though they would be important for a comprehensive data analysis
summarydata <-
all_dat %>%
select(
id, ahiTotal, cesdTotal, # ID & questionnaire scores
sex, age, educ, income # demographic variables
) Look at the Data
Have a look at the types of data:
Rows: 992
Columns: 7
$ id <dbl> 1, 1, 2, 2, 2, 2, 2, 2, 3, 3, 4, 4, 4, 4, 5, 5, 5, 5, 5, 6, …
$ ahiTotal <dbl> 63, 73, 73, 89, 89, 93, 80, 77, 77, 85, 60, 67, 56, 61, 41, …
$ cesdTotal <dbl> 14, 6, 7, 10, 13, 8, 15, 12, 3, 5, 31, 31, 41, 35, 27, 32, 2…
$ sex <dbl> 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 1, …
$ age <dbl> 35, 35, 59, 59, 59, 59, 59, 59, 51, 51, 50, 50, 50, 50, 58, …
$ educ <dbl> 5, 5, 1, 1, 1, 1, 1, 1, 4, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, …
$ income <dbl> 3, 3, 1, 1, 1, 1, 1, 1, 3, 3, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, …What do you see?
All variables are loaded as numeric. However, are all of those numeric?
sex, educ and income don’t seem to really be numbers but factors with individual categories (factor levels)!
We should convert these data to factor. Checking and adjusting data types (as part of data wrangling) will be important for plotting and analyzing the data, you might otherwise get strange/wrong results!
Activity 2: Transform to factor
Copy and run the below code to change the categories to factors.
- If you mutate a new column with the same name as the old one, it will overwrite the column.
Rows: 992
Columns: 7
$ id <dbl> 1, 1, 2, 2, 2, 2, 2, 2, 3, 3, 4, 4, 4, 4, 5, 5, 5, 5, 5, 6, …
$ ahiTotal <dbl> 63, 73, 73, 89, 89, 93, 80, 77, 77, 85, 60, 67, 56, 61, 41, …
$ cesdTotal <dbl> 14, 6, 7, 10, 13, 8, 15, 12, 3, 5, 31, 31, 41, 35, 27, 32, 2…
$ sex <fct> 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 1, …
$ age <dbl> 35, 35, 59, 59, 59, 59, 59, 59, 51, 51, 50, 50, 50, 50, 58, …
$ educ <fct> 5, 5, 1, 1, 1, 1, 1, 1, 4, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, …
$ income <fct> 3, 3, 1, 1, 1, 1, 1, 1, 3, 3, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, …Activity 2: Transform to factor
Rows: 992
Columns: 7
$ id <dbl> 1, 1, 2, 2, 2, 2, 2, 2, 3, 3, 4, 4, 4, 4, 5, 5, 5, 5, 5, 6, …
$ ahiTotal <dbl> 63, 73, 73, 89, 89, 93, 80, 77, 77, 85, 60, 67, 56, 61, 41, …
$ cesdTotal <dbl> 14, 6, 7, 10, 13, 8, 15, 12, 3, 5, 31, 31, 41, 35, 27, 32, 2…
$ sex <fct> 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 1, …
$ age <dbl> 35, 35, 59, 59, 59, 59, 59, 59, 51, 51, 50, 50, 50, 50, 58, …
$ educ <fct> 5, 5, 1, 1, 1, 1, 1, 1, 4, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, …
$ income <fct> 3, 3, 1, 1, 1, 1, 1, 1, 3, 3, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, …- At first glance, the data look the same. But the 1s and 2s in
sexare now e.g. not treated as numbers anymore, but as factor levels (categories). This e.g. changes how the data behave in different analyses, or plotting (continuous vs. categorical data).
Set labels
A simple change to a factor is not always helpful. We still don’t know what a 1 in sex or a 5 in educ stands for:
sex: 1 = female, 2 = maleeduc: 1 = no graduation, 2 = school graduation, 3 = vocational training, 4 = bachelor’s degree, 5 = post graduateincome: 1 = low, 2 = middle, 3 = high
Set labels
match_case() allows us to label our numeric data with more human-readable descriptions. if_else() is a useful shorthand for cases where we only have two categories.
summarydata2 <-
summarydata %>%
mutate(
sex = if_else(sex == 1, "female", "male") %>% as_factor(),
educ = educ %>%
case_match(
1 ~ "no graduation",
2 ~ "school graduation",
3 ~ "vocational training",
4 ~ "bachelor's degree",
5 ~ "post grad"
) %>%
as_factor(), # need to transform to factor in the end
income = income %>%
case_match(1 ~ "low", 2 ~ "middle", 3 ~ "high") %>%
as_factor()
)Set labels
[1] post grad no graduation bachelor's degree
[4] school graduation vocational training
5 Levels: post grad no graduation bachelor's degree ... vocational trainingFactor is now ordered by occurrence in data! :( (E.g., the order on the axis of a plot would be wrong.)
Set factor order (levels)
Using factor(), we can explicitly order the categories using the argument levels.
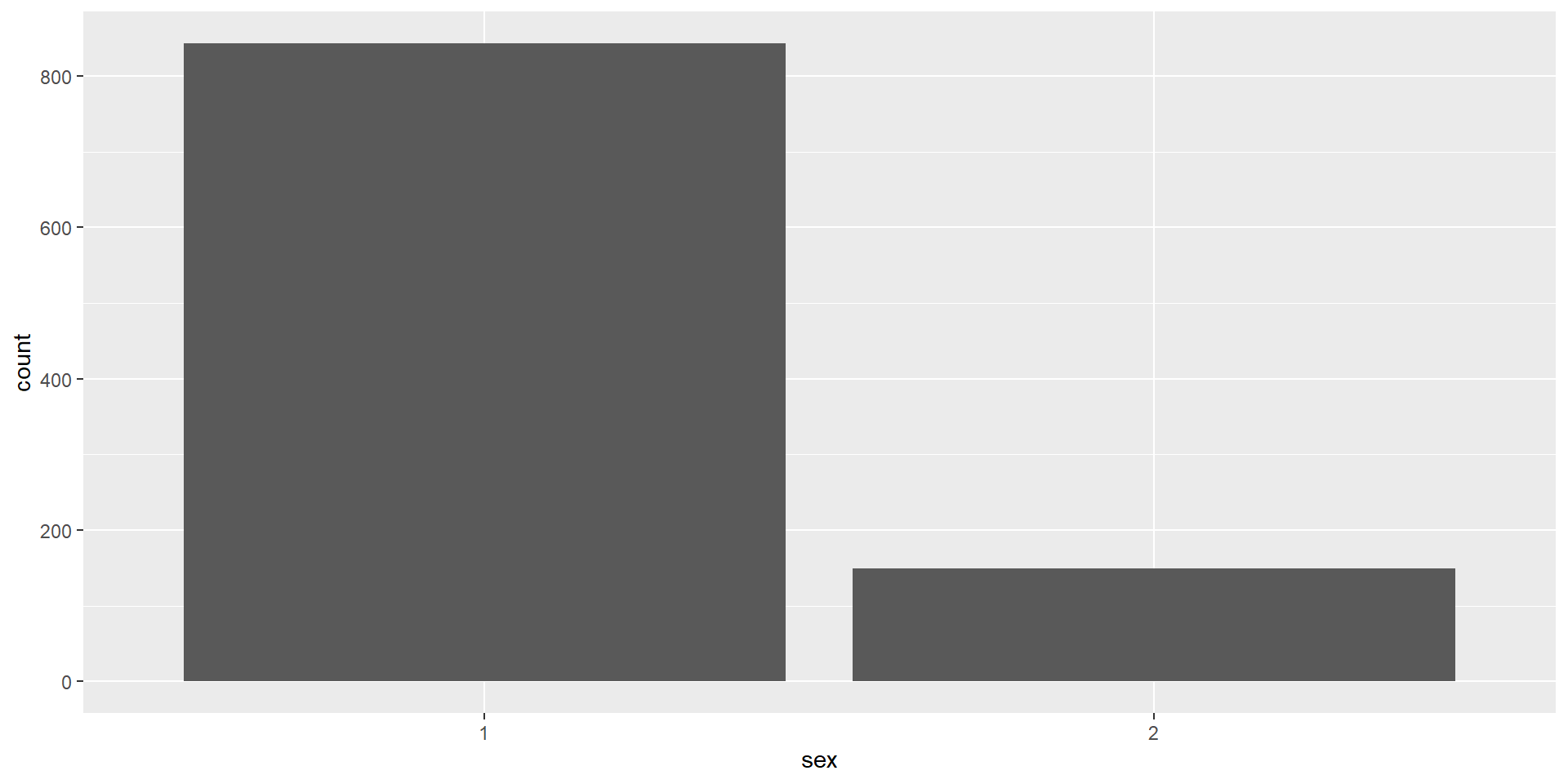
Activity 3: Barplot
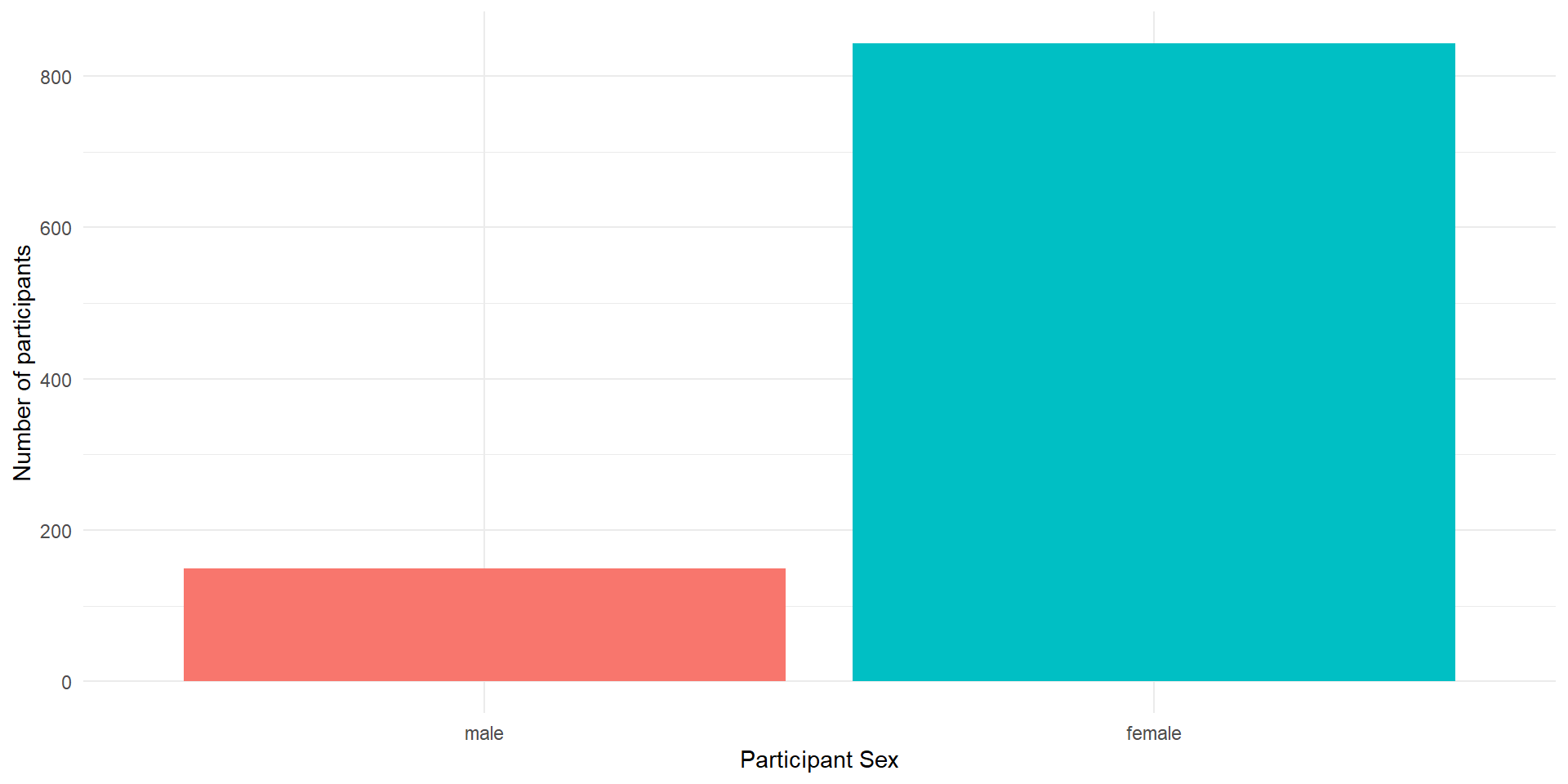
A bar plot is a plot that shows counts of categorical data (factors), where the height of each bar represents the count of that variable.
We will plot male and female participants.
The First Layer
The first line (or layer) sets up the base of the graph: the data to use and the aesthetics (
aes()) (what will go on the x and y axis, how the plot will be grouped).aes()includes anything that is directly related to your data: e.g., what goes on the x and y axis, or whether the plot should be grouped by a variable in your data.We can provide
xandyas arguments, however, in a bar plot, the count per category is calculated automatically, so we don’t need to put anything on the y-axis ourselves.
The Second Layer
The next layer adds a geom or a shape. In this case we use geom_bar() as we want to draw a bar plot.
- Note that we are adding layers, using a
+between layers. This is a very important difference between pipes and visualization.
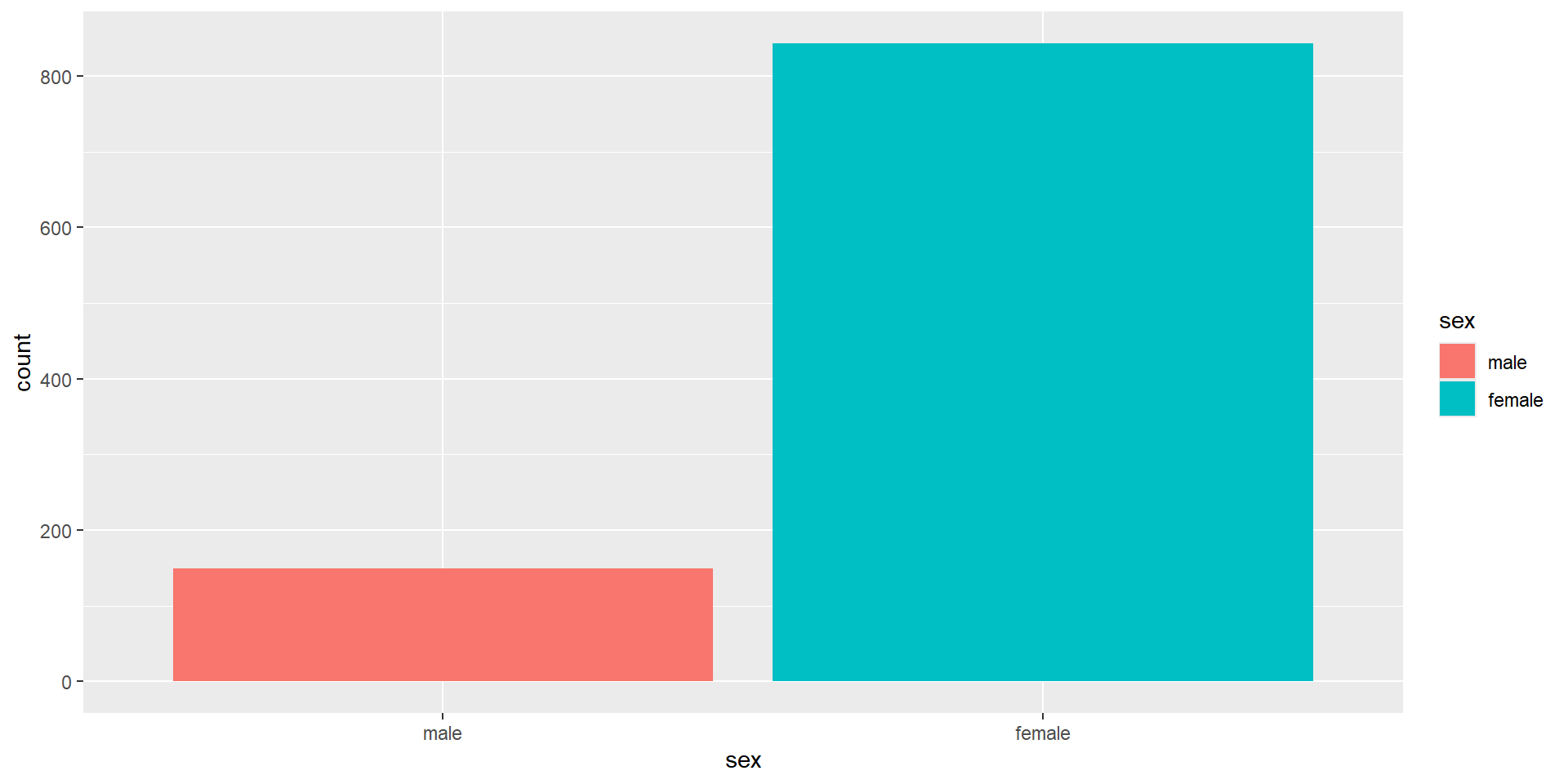
The Second Layer with color
Adding
fillto the first layer will separate the data into each level of the grouping variable and fill it with a different color. Note thatfillcolors the inside of the bar, whilecolourcolors the bar’s outlines.We can get rid of the (in this case redundant legend) with
show.legend = FALSE.
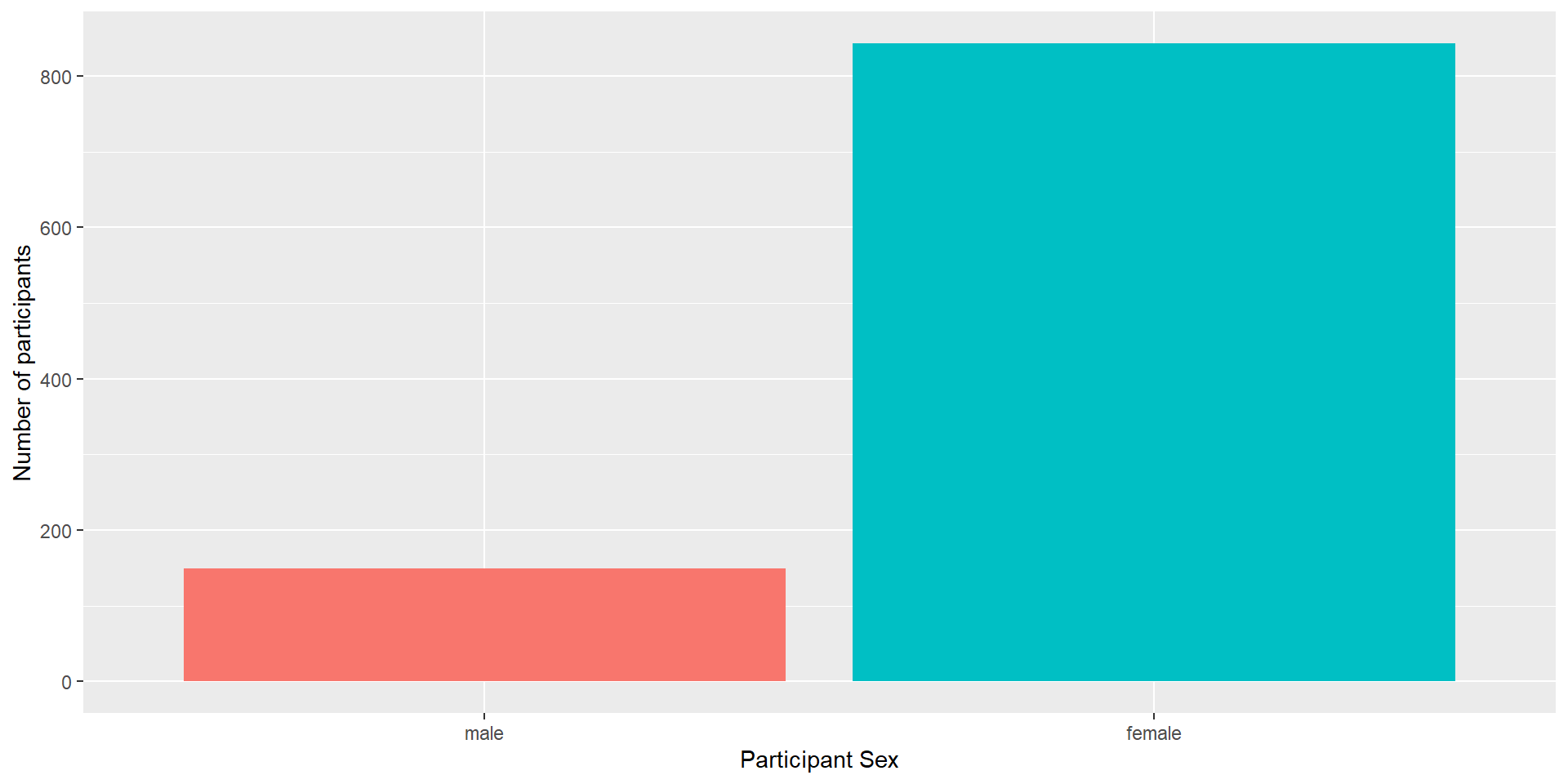
The Next Layers - Improving the Plot
We might want to make the plot a bit prettier and easier to read. What would you improve?
We could add better axis labels, and custom colors. We can do so with the functions scale_x_discrete() and scale_y_continuous(), which adjust the x and y axes.
Both functions can change several aspects of our axes; here, we use the argument name to set a new axis name.
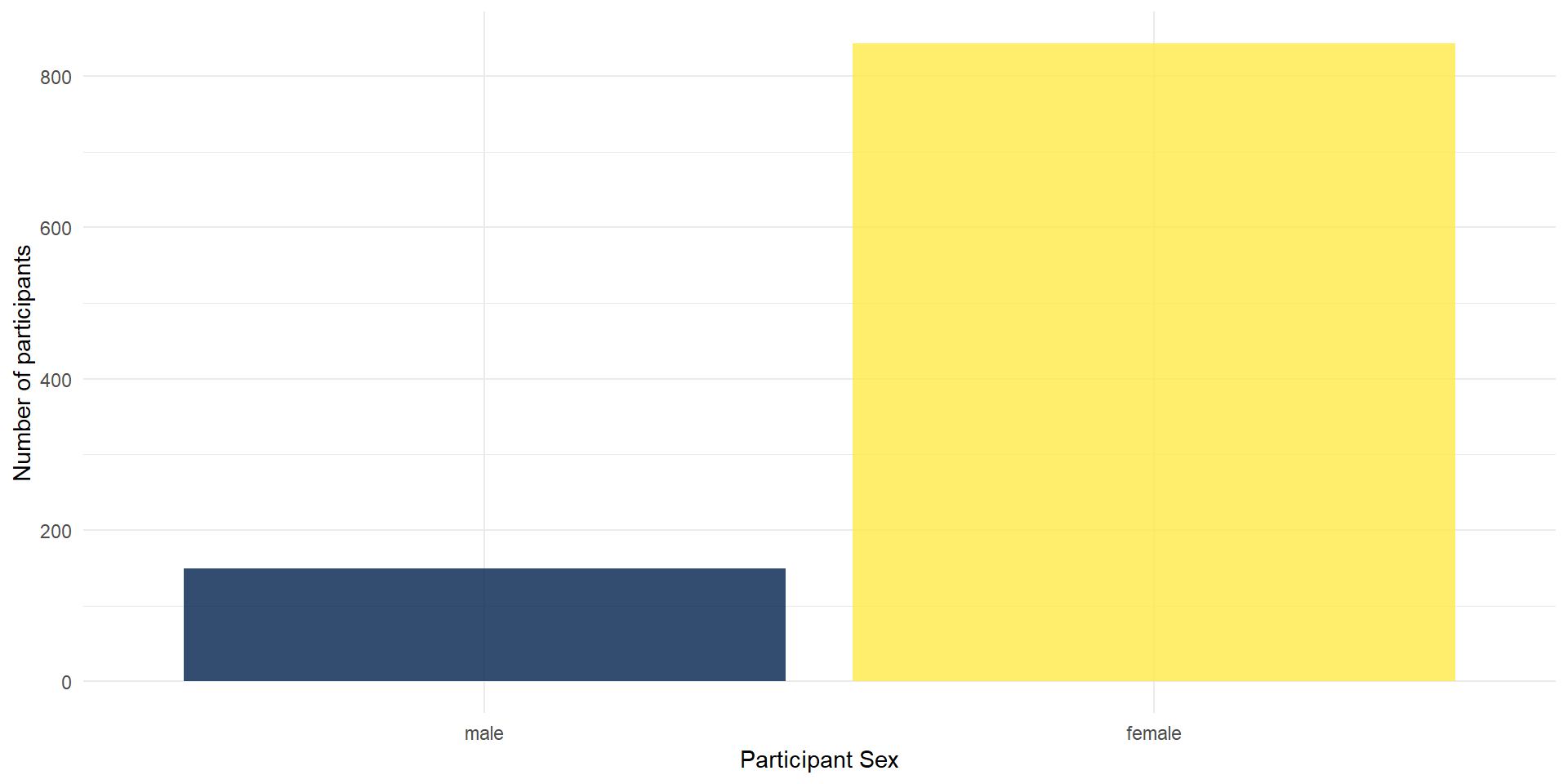
Themes: Changing the Appearance
There are a number of built-in themes that you can use to change the appearance (background, whether axes are shown etc.), but you can also tweak the themes further manually.
We will now change the default theme to theme_minimal(), but you can also try other themes (just type “theme_” and see what the autocomplete brings up).
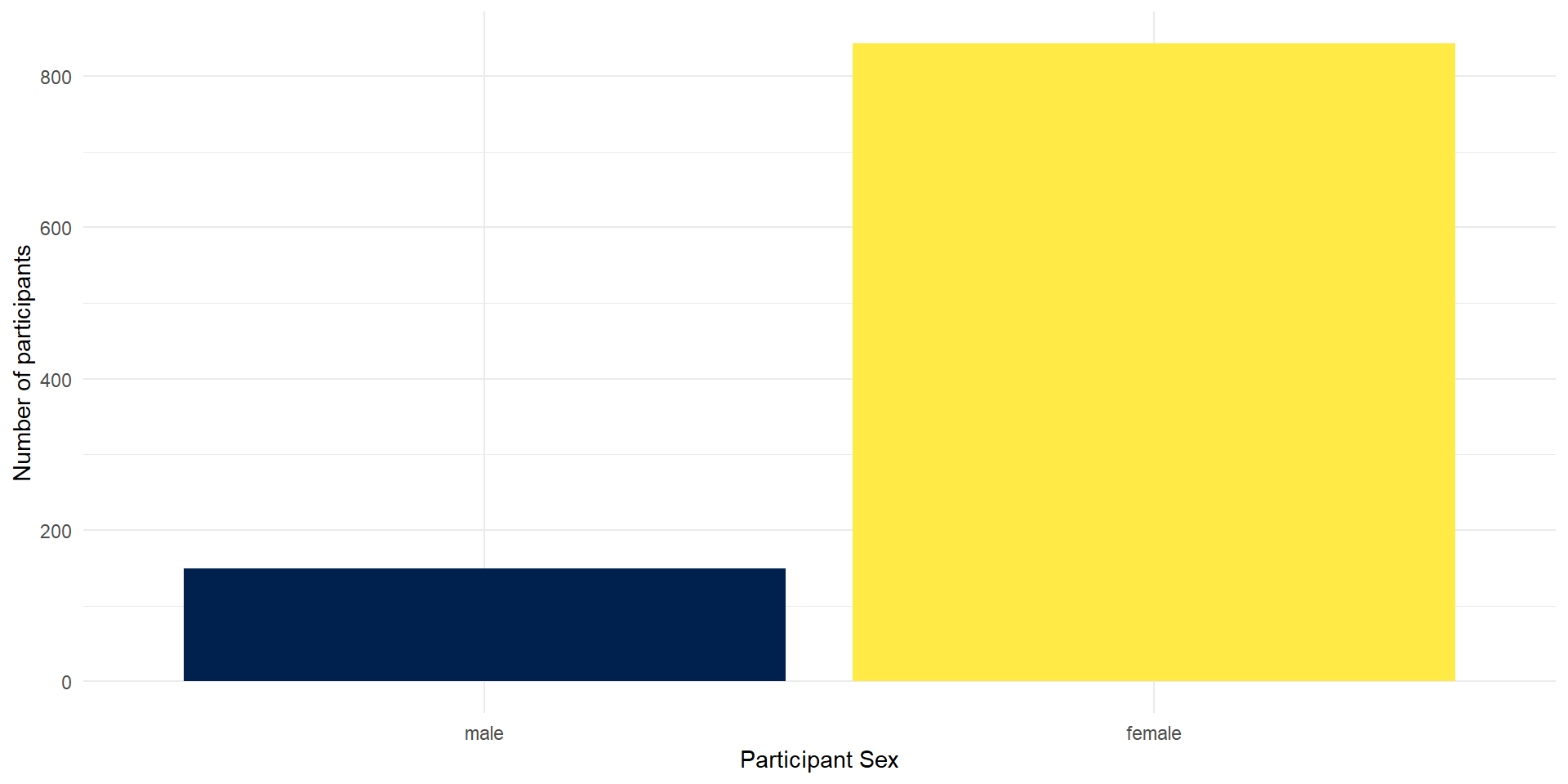
Colors
There are various ways to change the colors of the bars. You can manually indicate the colors you want to use but you can also easily use pre-determined color palettes that are already checked for color-blind friendliness.
A popular palette is viridis. We can simply add a function/layer to your ggplot named scale_fill_viridis_d() (d for discrete). The function has an option parameter that takes 5 different values (A - E).
- Run the code below. Try changing the option to either A, B, C or D and see which one you like!
Transparency
You can also add transparency to your plot, which can be helpful if you plot several layers of data that overlap.
To do so, you can simply add alpha to the geom_bar() - try changing the value of alpha (between 0 and 1):
Grouped Plots
Imagine that you have several factors that you want to use to group your data, such as gender and income. In this case, you could use a grouped bar plot:
ggplot(summarydata2, aes(x = sex, fill = income)) +
geom_bar(
# the default are stacked bars; we use "dodge" to put them side by side
position = "dodge",
alpha = .8
) +
scale_x_discrete(name = "Participant Sex") +
scale_y_continuous(name = "Number of participants") +
theme_minimal() +
scale_fill_viridis_d(option = "E")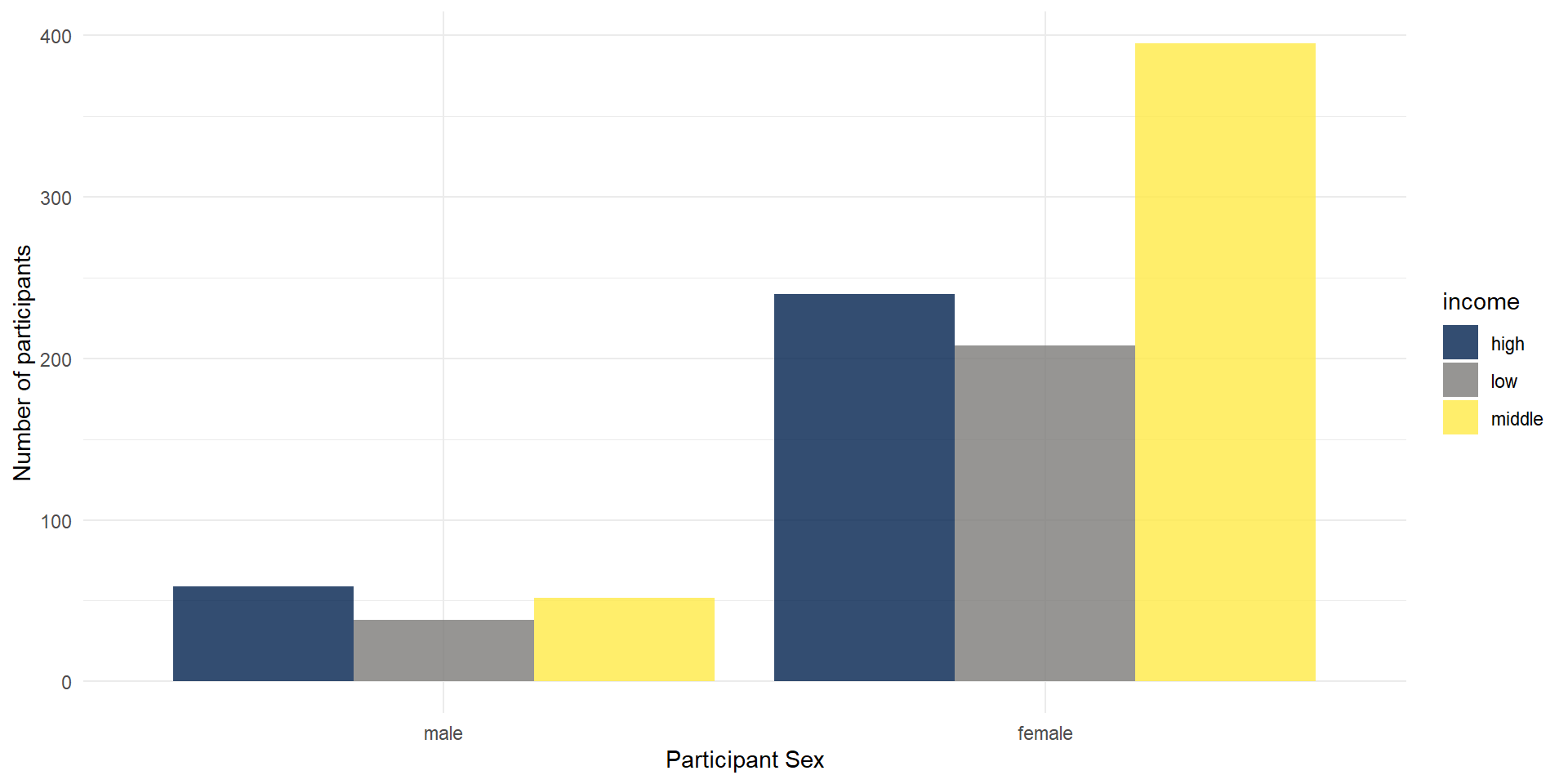
Facetting
You could also use facets to divide your data visualizations into several subplots: facet_wrap for one variable.
ggplot(summarydata2, aes(x = sex, fill = sex)) +
geom_bar(show.legend = FALSE, alpha = .8) +
scale_x_discrete(name = "Participant Sex") +
scale_y_continuous(name = "Number of participants") +
theme_minimal() +
scale_fill_viridis_d(option = "E") +
facet_wrap(vars(income)) # here, you need to use vars() around variable names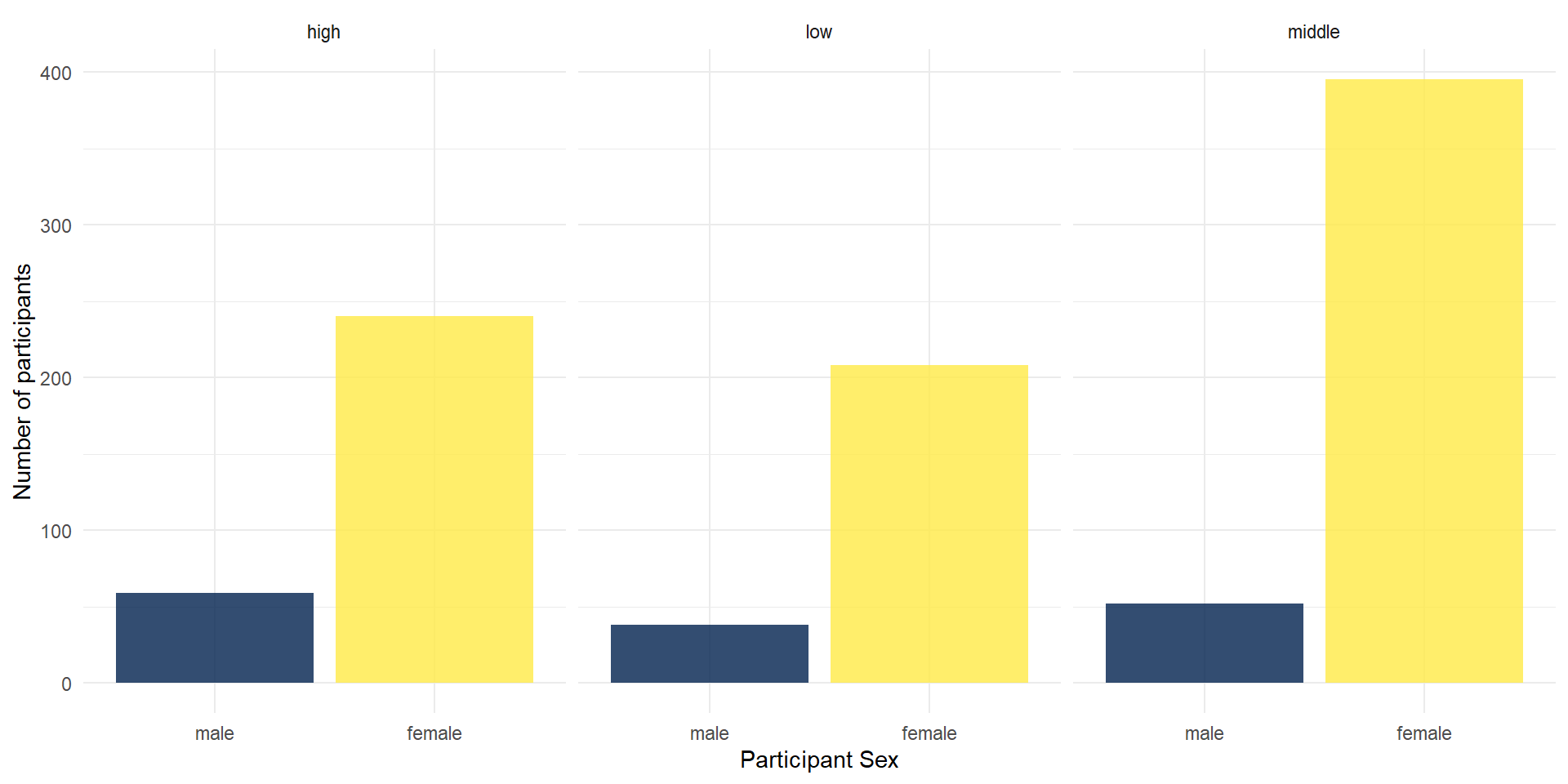
Facetting 2
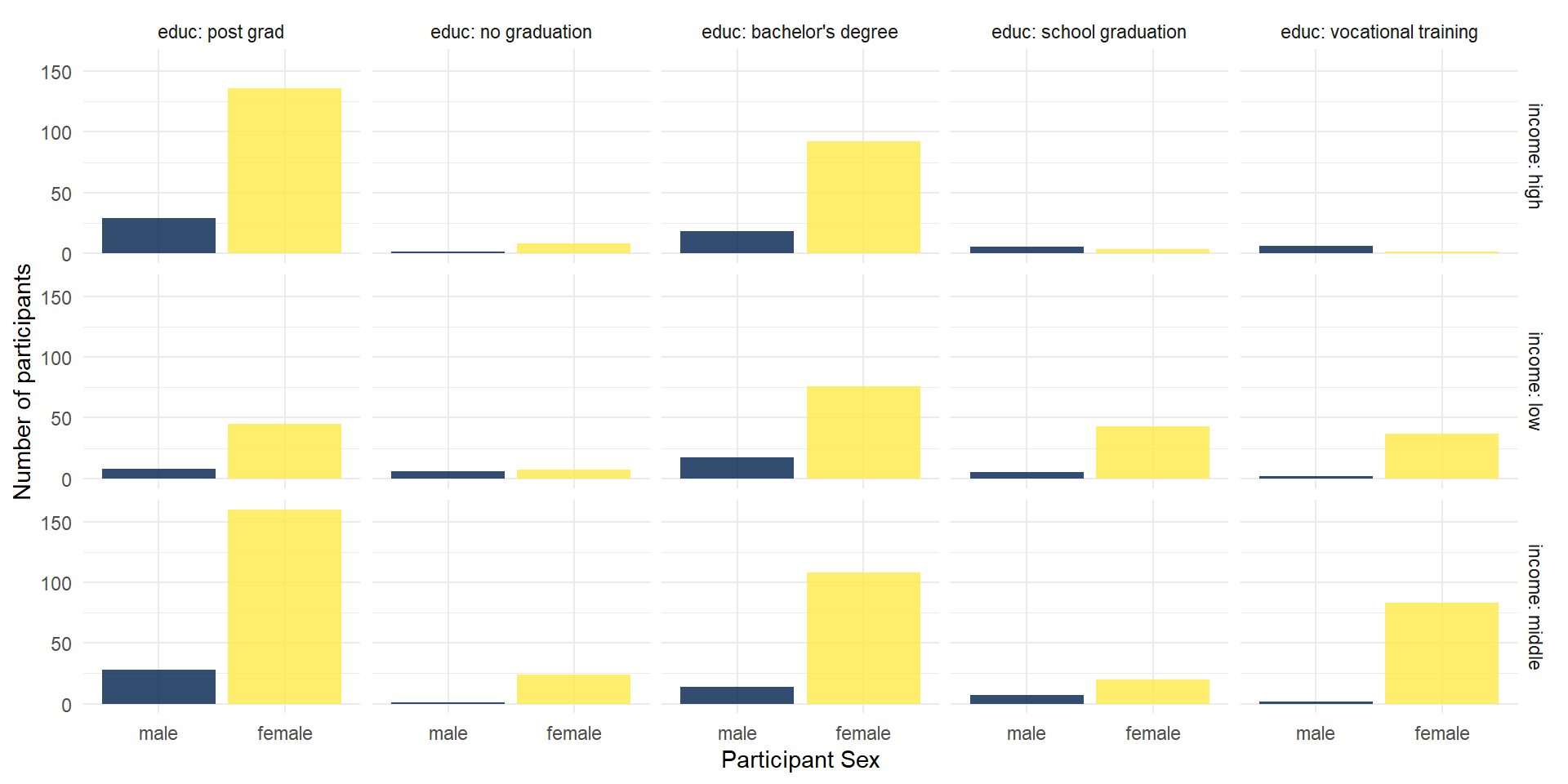
You could also use facets to divide your data visualizations into several subplots: facet_grid for a matrix of (combinations of) two variables.
ggplot(summarydata2, aes(x = sex, fill = sex)) +
geom_bar(show.legend = FALSE, alpha = .8) +
scale_x_discrete(name = "Participant Sex") +
scale_y_continuous(name = "Number of participants") +
theme_minimal() +
scale_fill_viridis_d(option = "E") +
facet_grid(
rows = vars(income),
cols = vars(educ),
labeller = "label_both" # this adds the variable name into the facet legends
)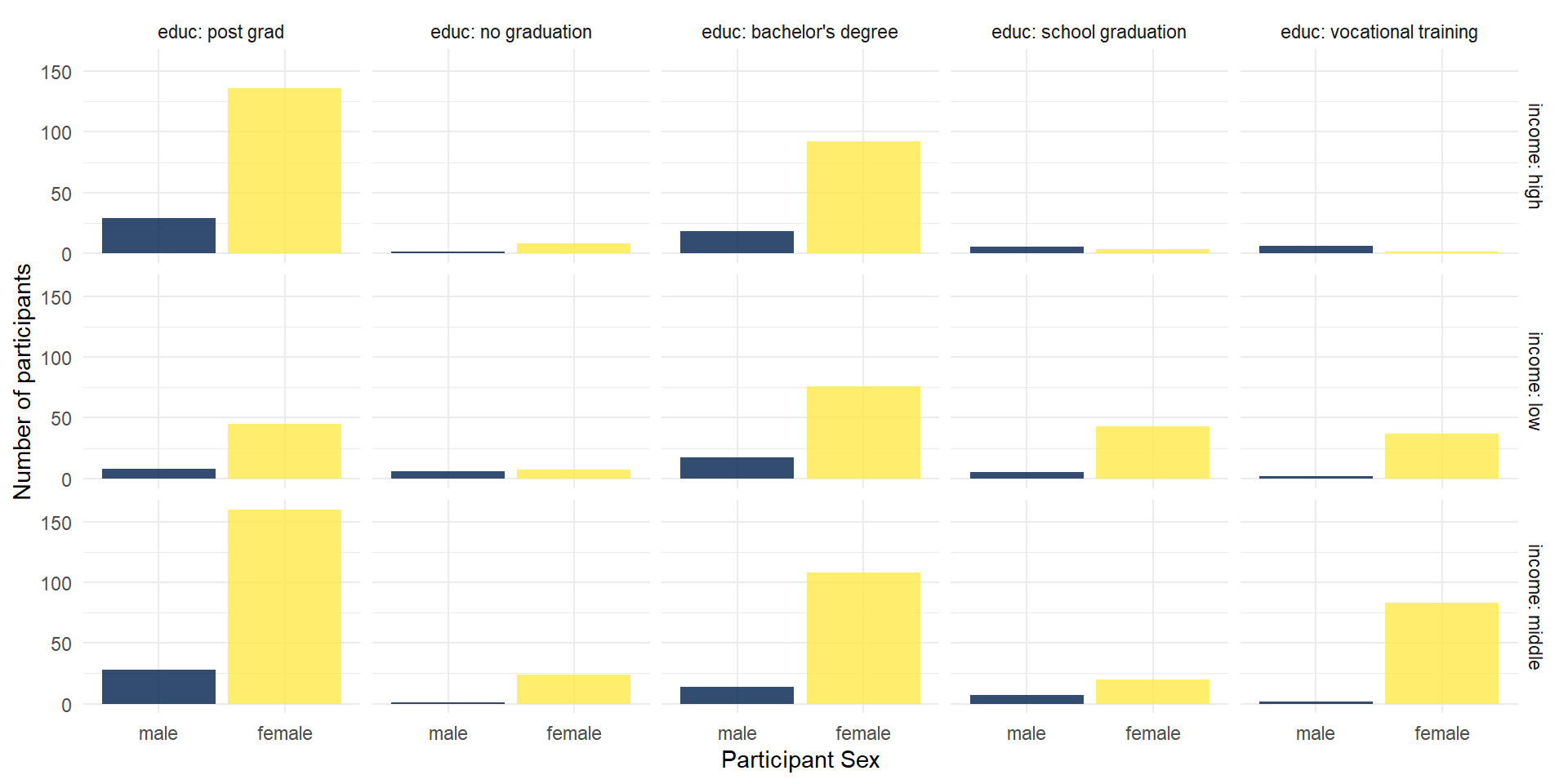
A closer look
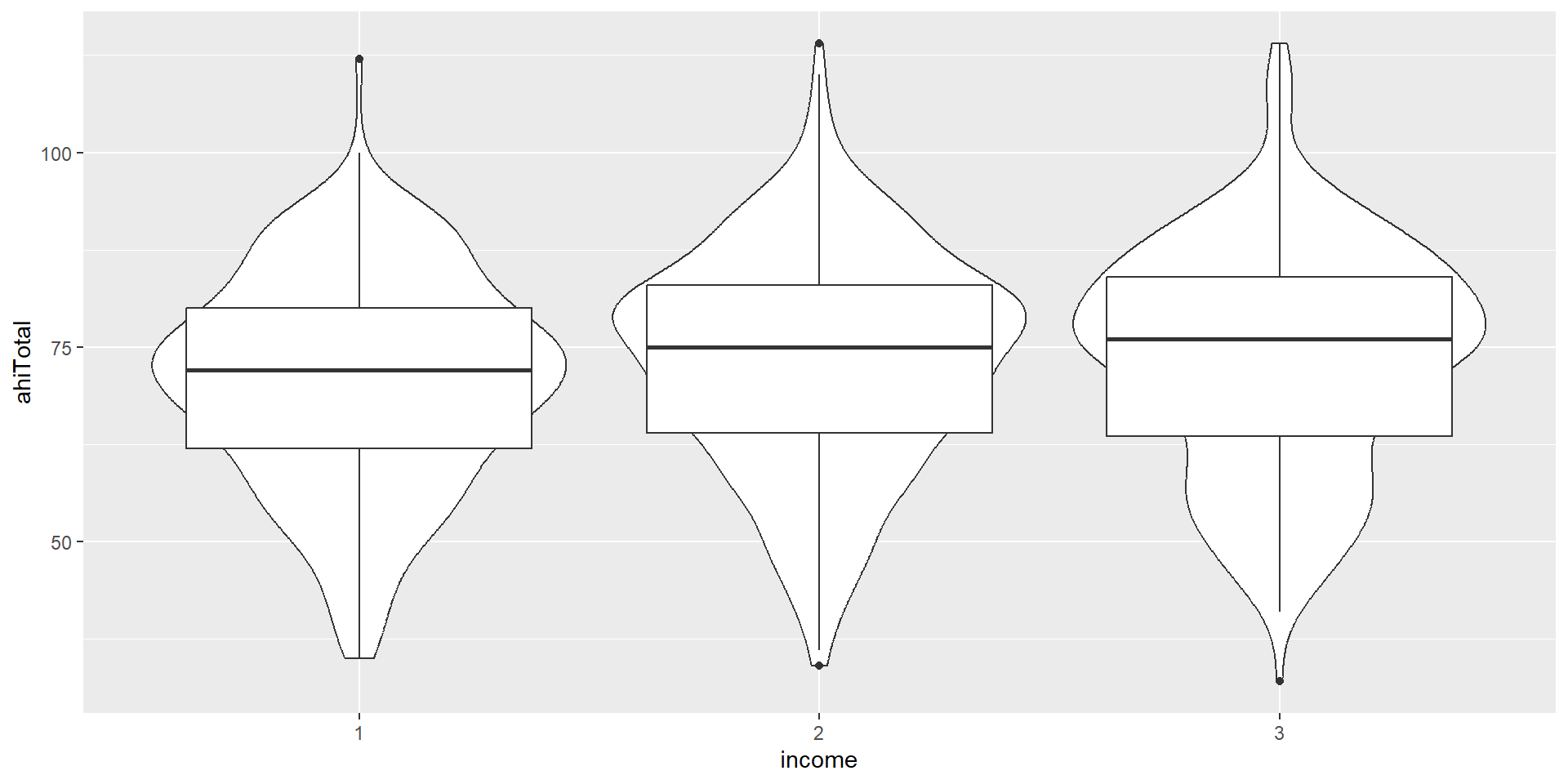
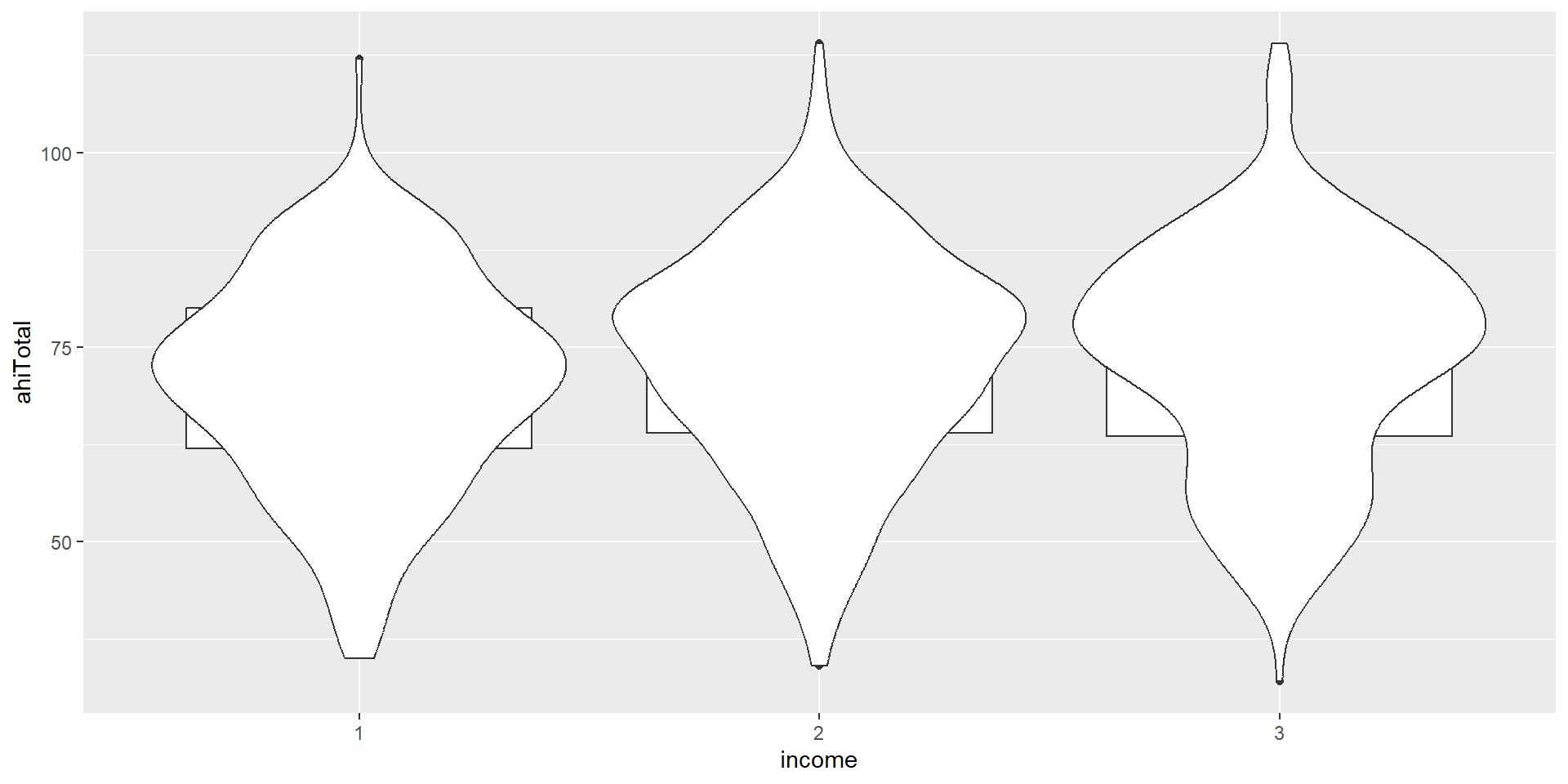
Activity 4: The Violin-Boxplot
There are a number of different figure types you can plot with the different geoms, e.g. geom_point(), geom_histogram(), geom_line()…
We now want to plot a form of a boxplot that becomes more popular: The violin-boxplot (which combines, i.e. overlays, the violin plot with a boxplot).
Violin-Boxplot
Let’s look at the code. How does the code differ from the one for the barplot above?
ggplot(summarydata1, aes(x = income,
y = ahiTotal, # new variable!
fill = income)) +
geom_violin(trim = FALSE, # smooth on edges
alpha = .4) +
geom_boxplot(width = .2, # small boxplot contained in violin
alpha = .7) +
scale_x_discrete(
name = "Income",
# set new labels
labels = c("Below Average", "Average", "Above Average")) +
scale_y_continuous(name = "Authentic Happiness Inventory Score")+
theme_minimal() +
# no need to switch of axis for every geom individually
theme(legend.position = "none") +
scale_fill_viridis_d()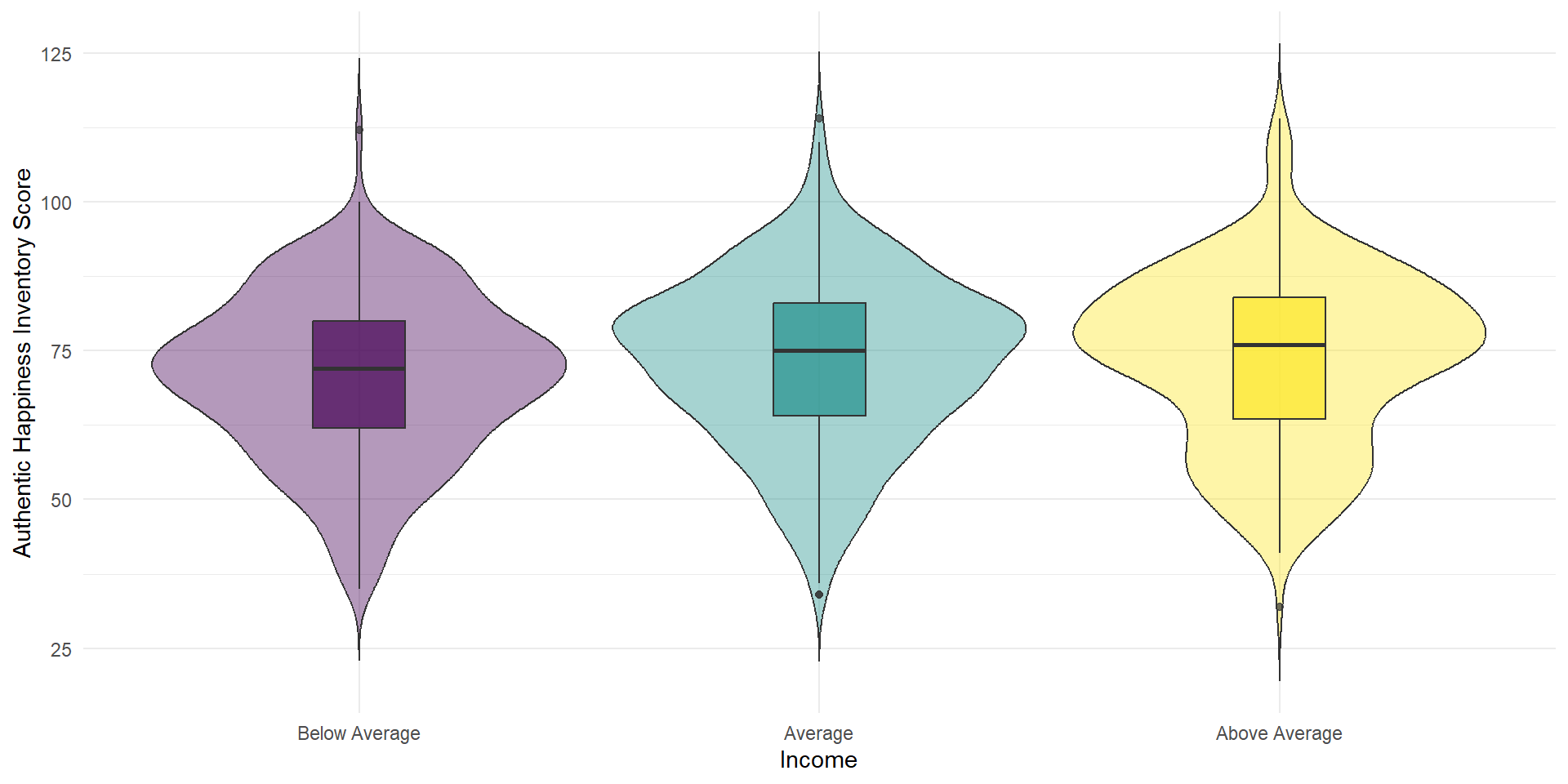
Layer Order
The order of layers is crucial, as the plot will be built up in that order (later layers on top):
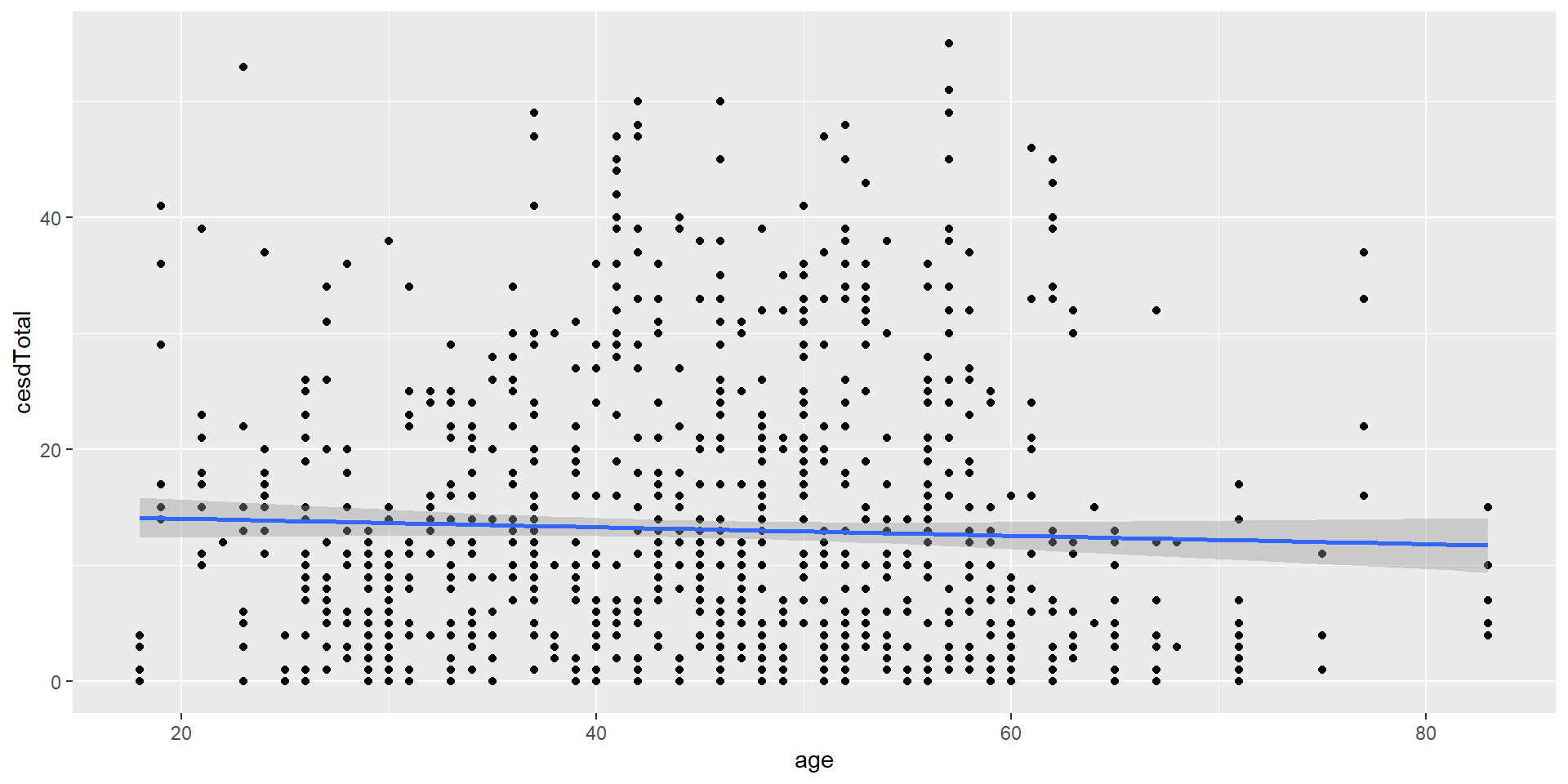
Scatterplot
If we have continuous data of two variables, we often want to make a scatter plot:
Saving your Figures
You can use ggsave() to save your plots. If you don’t tell ggsave() which plot you want to save, by default it will save the last plot you created.
You just have to enter the name of the file to be saved (in your working directory) like this:
Check whether indeed the last plot was saved!
You can also specify the dimensions of your plot to be saved:
or
Saving your Figures 2
You can also assign the plot to a variable in your environment and then tell ggsave() which object to save. This is a bit safer.
Run the code for the violin-boxplot again and save the plot in an object called viobox. You’d then have to explicitly tell ggsave() to save the object viobox:
viobox <-
ggplot(summarydata1, aes(x = income, y = ahiTotal, fill = income)) +
geom_violin(trim = FALSE, alpha = .4) +
geom_boxplot(width = .2, alpha = .7) +
scale_x_discrete(
name = "Income",
labels = c("Below Average", "Average", "Above Average")) +
scale_y_continuous(name = "Authentic Happiness Inventory Score")+
theme_minimal() +
theme(legend.position = "none") +
scale_fill_viridis_d()
ggsave("violin-boxplot-stored.png", plot = viobox)Thanks!
Check out Chapter 13 of QuantFun for further exercises and tips for data visualization!
The free book R for data science was written by (some of) the creators of the tidyverse, and includes lots of plotting and data science exercises. (For more advanced plotting, check the ggplot2 book.)
Also keep in mind: Coding is a lot of googling things!
You can also check out the R Graph Gallery for code for different types of plots.